military_clean %>%
filter(
if_all(.cols = -Country,
.fns = ~ is.na(.x)
),
!is.na(Country)
) %>%
pull(Country)Extending Joins, Factors, Clean Variable Names
Thursday, October 17
Today we will…
- Debrief PA 4
- Describe the Code
- Clean Column Names
- Debrief Lab 3 & Challenge 3
- Common Themes
- Package Lifecycle Stages
- Expectations for Tools Used
- Reminder about Lab 3 Peer Review
- New Material
- Extensions to Relational Data
- Factors with
forcats
- Lab 4: Childcare Costs in California
Practice Activity 4

Take 3-minutes to…
Write down in plain language what each line of this code is doing.
janitor Package

Image by Allison Horst
Clean Variable Names with janitor
Data from external sources likely has variable names not ideally formatted for R.
Names may…
- contain spaces.
- start with numbers.
- start with a mix of capital and lower case letters.
Clean Variable Names with janitor
The janitor package converts all variable names in a dataset to snake_case.
Names will…
- start with a lower case letter.
- have spaces filled in with
_.
Lab 3 Common Themes
Q1: The
tidyversepackage automatically loadsggplot2,dplyr,readr, etc. – do not load these twice!Q3: Where did these data come from? How were they collected? What is the context of these data?
- Challenge 3: When reaching a conclusion with the hypothesis test, what does Question 3 refer to?
Saving an f*$# load of objects
- Not outputting the results
Lab 3 Common Themes
- Not using
.xto specify where the.colsinput should go will go awry when there are multiple function inputs. - Using named arguments (e.g.,
.cols =,.fns =) makes your code more readable and is part of the code formatting guidelines for this class.
- Think about “efficient” ways to do things
- Q5: Are you using the same function
across()multiple columns? - Q6: Can you calculate multiple summary statistics in one pipeline?
- Q10-12: Is there a way you can get both the max and min in one pipeline?
- Q5: Are you using the same function
Lifecycle Stages
Lifceycle Stages
As packages get updated, the functions and function arguments included in those packages will change.
- The accepted syntax for a function may change.
- A function/functionality may disappear.
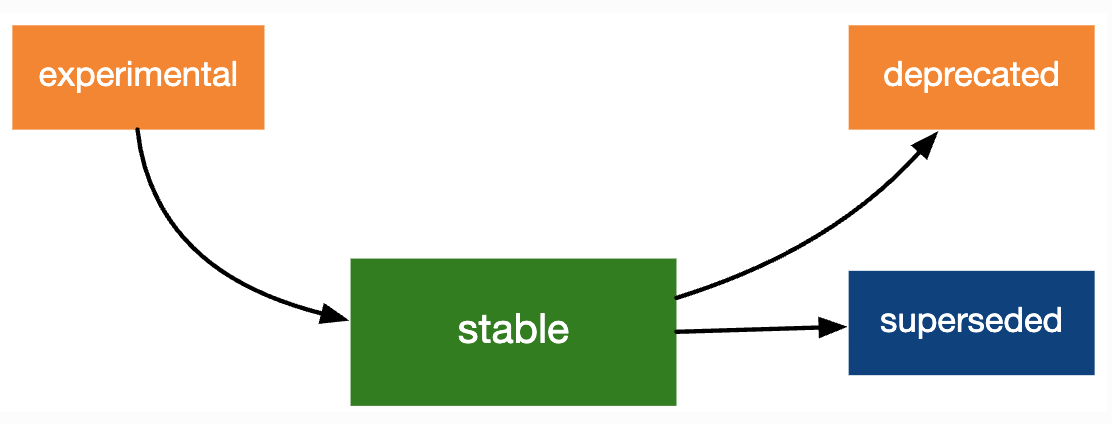
Learn more about lifecycle stages of packages, functions, function arguments in R.
Lifceycle Stages

Deprecated Functions
A deprecated functionality has a better alternative available and is scheduled for removal.
- You get a warning telling you what to use instead.
Warning: Using `across()` in `filter()` was deprecated in dplyr 1.0.8.
ℹ Please use `if_any()` or `if_all()` instead.# A tibble: 18 × 35
Country Notes `Reporting year` `1988` `1989` `1990` `1991` `1992` `1993`
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 Africa <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
2 North Africa <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
3 Sub-Saharan <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
4 Americas <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
5 Central Ame… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
6 North Ameri… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
7 South Ameri… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
8 Asia & Ocea… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
9 Central Asia <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
10 East Asia <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
11 South Asia <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
12 South-East … <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
13 Oceania <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
14 Europe <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
15 Central Eur… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
16 Eastern Eur… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
17 Western Eur… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
18 Middle East <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
# ℹ 26 more variables: `1994` <chr>, `1995` <chr>, `1996` <chr>, `1997` <chr>,
# `1998` <chr>, `1999` <chr>, `2000` <chr>, `2001` <chr>, `2002` <chr>,
# `2003` <chr>, `2004` <chr>, `2005` <chr>, `2006` <chr>, `2007` <chr>,
# `2008` <chr>, `2009` <chr>, `2010` <chr>, `2011` <chr>, `2012` <chr>,
# `2013` <chr>, `2014` <chr>, `2015` <chr>, `2016` <chr>, `2017` <chr>,
# `2018` <chr>, `2019` <chr>Deprecated Functions
You should not use deprecated functions!
Instead, we use…
# A tibble: 18 × 35
Country Notes `Reporting year` `1988` `1989` `1990` `1991` `1992` `1993`
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 Africa <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
2 North Africa <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
3 Sub-Saharan <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
4 Americas <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
5 Central Ame… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
6 North Ameri… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
7 South Ameri… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
8 Asia & Ocea… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
9 Central Asia <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
10 East Asia <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
11 South Asia <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
12 South-East … <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
13 Oceania <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
14 Europe <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
15 Central Eur… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
16 Eastern Eur… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
17 Western Eur… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
18 Middle East <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
# ℹ 26 more variables: `1994` <chr>, `1995` <chr>, `1996` <chr>, `1997` <chr>,
# `1998` <chr>, `1999` <chr>, `2000` <chr>, `2001` <chr>, `2002` <chr>,
# `2003` <chr>, `2004` <chr>, `2005` <chr>, `2006` <chr>, `2007` <chr>,
# `2008` <chr>, `2009` <chr>, `2010` <chr>, `2011` <chr>, `2012` <chr>,
# `2013` <chr>, `2014` <chr>, `2015` <chr>, `2016` <chr>, `2017` <chr>,
# `2018` <chr>, `2019` <chr>Superceded Functions
A superseded functionality has a better alternative, but is not going away.
- This is a softer alternative to deprecation.
- A superseded function will not give a warning (since there’s no risk if you keep using it), but the documentation will give you a recommendation for what to use instead.
What is my job?
Teaching you stuff
(Thoughtfully) choosing what to teach and how to teach it.
Assessing what you’ve learned
What do you understand about the tools I’ve taught you?
This is not the same as assessing if you figured out a way to accomplish a given task.
Don’t Forget to Complete Your Lab 3 Code Review
Make sure your feedback follows the code review guidelines.
Insert your review into the comment box!
Extensions to Relational Data
Relational Data
When we work with multiple tables of data, we say we are working with relational data.
- It is the relations, not just the individual datasets, that are important.
When we work with relational data, we rely on keys.
- A key uniquely identifies an observation in a dataset.
- A key allows us to relate datasets to each other
IMDb Movies Data

How can we find each director’s active years?
Joining Multiple Data Sets

| director_id | movie_id | first_name | last_name |
|---|---|---|---|
| 429 | 300229 | Andrew | Adamson |
| 2931 | 254943 | Darren | Aronofsky |
| 9247 | 124110 | Zach | Braff |
| 11652 | 10920 | James (I) | Cameron |
| 11652 | 333856 | James (I) | Cameron |
| 14927 | 192017 | Ron | Clements |
| 15092 | 109093 | Ethan | Coen |
| 15092 | 237431 | Ethan | Coen |
| 15093 | 109093 | Joel | Coen |
| 15093 | 237431 | Joel | Coen |
| 15901 | 130128 | Francis Ford | Coppola |
| 15906 | 194874 | Sofia | Coppola |
| 16816 | 350424 | Cameron | Crowe |
| 17810 | 297838 | Frank | Darabont |
| 22104 | 224842 | Clint | Eastwood |
| 24758 | 112290 | David | Fincher |
| 28395 | 46169 | Mel (I) | Gibson |
| 35573 | 18979 | Ron | Howard |
| 35838 | 257264 | John (I) | Hughes |
| 37872 | 300229 | Vicky | Jenson |
| 38746 | 238695 | Mike (I) | Judge |
| 41975 | 314965 | David | Koepp |
| 44291 | 17173 | John (I) | Landis |
| 46315 | 344203 | Jay | Levey |
| 48115 | 313459 | George | Lucas |
| 56332 | 192017 | John | Musker |
| 58201 | 30959 | Christopher | Nolan |
| 58201 | 210511 | Christopher | Nolan |
| 65940 | 111813 | Rob | Reiner |
| 66849 | 306032 | Guy | Ritchie |
| 68161 | 116907 | Herbert (I) | Ross |
| 74758 | 238072 | Steven | Soderbergh |
| 76524 | 167324 | Oliver (I) | Stone |
| 78273 | 176711 | Quentin | Tarantino |
| 78273 | 176712 | Quentin | Tarantino |
| 78273 | 267038 | Quentin | Tarantino |
| 78273 | 276217 | Quentin | Tarantino |
| 82525 | 147603 | Paul (I) | Verhoeven |
| 83616 | 207992 | Andy | Wachowski |
| 83617 | 207992 | Larry | Wachowski |
| 88802 | 256630 | Unknown | Director |
| director_id | movie_id | first_name | last_name | movie_name | year | rank |
|---|---|---|---|---|---|---|
| 429 | 300229 | Andrew | Adamson | Shrek | 2001 | 8.1 |
| 2931 | 254943 | Darren | Aronofsky | Pi | 1998 | 7.5 |
| 9247 | 124110 | Zach | Braff | Garden State | 2004 | 8.3 |
| 11652 | 10920 | James (I) | Cameron | Aliens | 1986 | 8.2 |
| 11652 | 333856 | James (I) | Cameron | Titanic | 1997 | 6.9 |
| 14927 | 192017 | Ron | Clements | Little Mermaid, The | 1989 | 7.3 |
| 15092 | 109093 | Ethan | Coen | Fargo | 1996 | 8.2 |
| 15092 | 237431 | Ethan | Coen | O Brother, Where Art Thou? | 2000 | 7.8 |
| 15093 | 109093 | Joel | Coen | Fargo | 1996 | 8.2 |
| 15093 | 237431 | Joel | Coen | O Brother, Where Art Thou? | 2000 | 7.8 |
| 15901 | 130128 | Francis Ford | Coppola | Godfather, The | 1972 | 9.0 |
| 15906 | 194874 | Sofia | Coppola | Lost in Translation | 2003 | 8.0 |
| 16816 | 350424 | Cameron | Crowe | Vanilla Sky | 2001 | 6.9 |
| 17810 | 297838 | Frank | Darabont | Shawshank Redemption, The | 1994 | 9.0 |
| 22104 | 224842 | Clint | Eastwood | Mystic River | 2003 | 8.1 |
| 24758 | 112290 | David | Fincher | Fight Club | 1999 | 8.5 |
| 28395 | 46169 | Mel (I) | Gibson | Braveheart | 1995 | 8.3 |
| 35573 | 18979 | Ron | Howard | Apollo 13 | 1995 | 7.5 |
| 35838 | 257264 | John (I) | Hughes | Planes, Trains & Automobiles | 1987 | 7.2 |
| 37872 | 300229 | Vicky | Jenson | Shrek | 2001 | 8.1 |
| 38746 | 238695 | Mike (I) | Judge | Office Space | 1999 | 7.6 |
| 41975 | 314965 | David | Koepp | Stir of Echoes | 1999 | 7.0 |
| 44291 | 17173 | John (I) | Landis | Animal House | 1978 | 7.5 |
| 46315 | 344203 | Jay | Levey | UHF | 1989 | 6.6 |
| 48115 | 313459 | George | Lucas | Star Wars | 1977 | 8.8 |
| 56332 | 192017 | John | Musker | Little Mermaid, The | 1989 | 7.3 |
| 58201 | 30959 | Christopher | Nolan | Batman Begins | 2005 | NA |
| 58201 | 210511 | Christopher | Nolan | Memento | 2000 | 8.7 |
| 65940 | 111813 | Rob | Reiner | Few Good Men, A | 1992 | 7.5 |
| 66849 | 306032 | Guy | Ritchie | Snatch. | 2000 | 7.9 |
| 68161 | 116907 | Herbert (I) | Ross | Footloose | 1984 | 5.8 |
| 74758 | 238072 | Steven | Soderbergh | Ocean's Eleven | 2001 | 7.5 |
| 76524 | 167324 | Oliver (I) | Stone | JFK | 1991 | 7.8 |
| 78273 | 176711 | Quentin | Tarantino | Kill Bill: Vol. 1 | 2003 | 8.4 |
| 78273 | 176712 | Quentin | Tarantino | Kill Bill: Vol. 2 | 2004 | 8.2 |
| 78273 | 267038 | Quentin | Tarantino | Pulp Fiction | 1994 | 8.7 |
| 78273 | 276217 | Quentin | Tarantino | Reservoir Dogs | 1992 | 8.3 |
| 82525 | 147603 | Paul (I) | Verhoeven | Hollow Man | 2000 | 5.3 |
| 83616 | 207992 | Andy | Wachowski | Matrix, The | 1999 | 8.5 |
| 83617 | 207992 | Larry | Wachowski | Matrix, The | 1999 | 8.5 |
| 88802 | 256630 | Unknown | Director | Pirates of the Caribbean | 2003 | NA |
Joining on Multiple Variables
Consider the rodent data from Lab 2.
- We want to add
species_idto the rodent measurements.
| genus | species | taxa | species_id |
|---|---|---|---|
| Dipodomys | merriami | Rodent | DM |
| Dipodomys | ordii | Rodent | DO |
| Perognathus | flavus | Rodent | PF |
| Chaetodipus | penicillatus | Rodent | PP |
| Peromyscus | eremicus | Rodent | PE |
| Onychomys | leucogaster | Rodent | OL |
| Reithrodontomys | megalotis | Rodent | RM |
| Dipodomys | spectabilis | Rodent | DS |
| Onychomys | torridus | Rodent | OT |
| Neotoma | albigula | Rodent | NL |
| Peromyscus | maniculatus | Rodent | PM |
| Sigmodon | hispidus | Rodent | SH |
| Reithrodontomys | fulvescens | Rodent | RF |
| Chaetodipus | baileyi | Rodent | PB |
| genus_name | species | sex | hindfoot_length | weight |
|---|---|---|---|---|
| Dipodomys | merriami | M | 35 | 40 |
| Dipodomys | merriami | M | 37 | 48 |
| Dipodomys | merriami | F | 34 | 29 |
| Dipodomys | merriami | F | 35 | 46 |
| Dipodomys | merriami | M | 35 | 36 |
| Dipodomys | ordii | F | 32 | 52 |
| Perognathus | flavus | M | 15 | 8 |
| Dipodomys | merriami | F | 36 | 35 |
| Perognathus | flavus | M | 12 | 7 |
| Dipodomys | merriami | F | 32 | 22 |
| Perognathus | flavus | M | 16 | 9 |
| Dipodomys | merriami | F | 34 | 42 |
| Perognathus | flavus | F | 14 | 8 |
| Dipodomys | merriami | F | 35 | 41 |
| Dipodomys | merriami | F | 37 | 37 |
| Dipodomys | merriami | F | 35 | 43 |
| Dipodomys | merriami | F | 35 | 41 |
| Dipodomys | merriami | F | 33 | 40 |
| Perognathus | flavus | F | 11 | 9 |
| Dipodomys | merriami | F | 35 | 45 |
| Chaetodipus | penicillatus | F | 20 | 15 |
| Dipodomys | merriami | M | 35 | 29 |
| Dipodomys | merriami | M | 35 | 39 |
| Dipodomys | merriami | F | 36 | 43 |
| Dipodomys | merriami | M | 38 | 46 |
| Dipodomys | merriami | M | 36 | 41 |
| Dipodomys | merriami | M | 36 | 41 |
| Dipodomys | merriami | M | 38 | 40 |
| Dipodomys | merriami | M | 37 | 45 |
| Dipodomys | merriami | F | 35 | 46 |
| Dipodomys | merriami | F | 35 | 40 |
| Dipodomys | merriami | F | 35 | 30 |
| Dipodomys | merriami | M | 35 | 39 |
| Dipodomys | merriami | M | 35 | 34 |
| Dipodomys | merriami | F | 37 | 42 |
| Dipodomys | merriami | M | 37 | 42 |
| Perognathus | flavus | F | 13 | 8 |
| Dipodomys | merriami | F | 37 | 31 |
| Dipodomys | merriami | F | 36 | 40 |
| Dipodomys | merriami | M | 36 | 37 |
| Dipodomys | merriami | M | 36 | 48 |
| Dipodomys | merriami | M | 37 | 42 |
| Dipodomys | merriami | F | 39 | 45 |
| Chaetodipus | penicillatus | F | 21 | 16 |
| Dipodomys | merriami | F | 36 | 36 |
| Dipodomys | merriami | M | 36 | 42 |
| Dipodomys | merriami | M | 36 | 44 |
| Dipodomys | merriami | F | 36 | 41 |
| Dipodomys | merriami | F | 36 | 40 |
| Dipodomys | merriami | M | 37 | 34 |
| Dipodomys | merriami | M | 33 | 40 |
| Dipodomys | merriami | M | 33 | 44 |
| Dipodomys | merriami | M | 37 | 44 |
| Dipodomys | merriami | M | 34 | 36 |
| Dipodomys | merriami | M | 35 | 33 |
| Dipodomys | merriami | F | 37 | 46 |
| Dipodomys | merriami | F | 34 | 35 |
| Dipodomys | merriami | M | 36 | 46 |
| Dipodomys | merriami | F | 33 | 37 |
| Dipodomys | merriami | M | 36 | 34 |
| Dipodomys | merriami | F | 36 | 45 |
| Perognathus | flavus | F | 15 | 7 |
| Dipodomys | merriami | M | 37 | 51 |
| Dipodomys | merriami | M | 35 | 39 |
| Dipodomys | merriami | M | 36 | 29 |
| Dipodomys | merriami | F | 32 | 48 |
| Dipodomys | merriami | M | 38 | 46 |
| Dipodomys | merriami | F | 37 | 41 |
| Dipodomys | merriami | M | 37 | 45 |
| Dipodomys | merriami | F | 35 | 42 |
| Dipodomys | merriami | F | 36 | 53 |
| Dipodomys | merriami | F | 35 | 49 |
| Dipodomys | merriami | F | 36 | 46 |
| Perognathus | flavus | F | 13 | 9 |
| Chaetodipus | penicillatus | F | 19 | 15 |
| Perognathus | flavus | M | 13 | 4 |
| Dipodomys | merriami | M | 36 | 48 |
| Dipodomys | merriami | M | 37 | 51 |
| Dipodomys | merriami | M | 38 | 50 |
| Dipodomys | merriami | M | 35 | 44 |
| Dipodomys | merriami | M | 25 | 44 |
| Dipodomys | merriami | M | 35 | 45 |
| Dipodomys | merriami | F | 37 | 45 |
| Peromyscus | eremicus | M | 20 | 19 |
| Dipodomys | merriami | F | 38 | 44 |
| Dipodomys | merriami | F | 36 | 42 |
| Dipodomys | merriami | M | 37 | 39 |
| Dipodomys | merriami | M | 37 | 47 |
| Dipodomys | merriami | M | 36 | 42 |
| Dipodomys | merriami | M | 36 | 49 |
| Dipodomys | merriami | M | 38 | 39 |
| Dipodomys | merriami | F | 36 | 43 |
| Dipodomys | merriami | M | 35 | 50 |
| Dipodomys | merriami | M | 36 | 41 |
| Dipodomys | merriami | M | 37 | 47 |
| Dipodomys | merriami | F | 36 | 37 |
| Dipodomys | merriami | M | 36 | 41 |
| Dipodomys | merriami | F | 36 | 36 |
| Dipodomys | merriami | M | 36 | 45 |
| Peromyscus | eremicus | M | 19 | 20 |
Join by species + genus
| genus_name | species | sex | hindfoot_length | weight | taxa | species_id |
|---|---|---|---|---|---|---|
| Dipodomys | merriami | M | 35 | 40 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 48 | Rodent | DM |
| Dipodomys | merriami | F | 34 | 29 | Rodent | DM |
| Dipodomys | merriami | F | 35 | 46 | Rodent | DM |
| Dipodomys | merriami | M | 35 | 36 | Rodent | DM |
| Dipodomys | ordii | F | 32 | 52 | Rodent | DO |
| Perognathus | flavus | M | 15 | 8 | Rodent | PF |
| Dipodomys | merriami | F | 36 | 35 | Rodent | DM |
| Perognathus | flavus | M | 12 | 7 | Rodent | PF |
| Dipodomys | merriami | F | 32 | 22 | Rodent | DM |
| Perognathus | flavus | M | 16 | 9 | Rodent | PF |
| Dipodomys | merriami | F | 34 | 42 | Rodent | DM |
| Perognathus | flavus | F | 14 | 8 | Rodent | PF |
| Dipodomys | merriami | F | 35 | 41 | Rodent | DM |
| Dipodomys | merriami | F | 37 | 37 | Rodent | DM |
| Dipodomys | merriami | F | 35 | 43 | Rodent | DM |
| Dipodomys | merriami | F | 35 | 41 | Rodent | DM |
| Dipodomys | merriami | F | 33 | 40 | Rodent | DM |
| Perognathus | flavus | F | 11 | 9 | Rodent | PF |
| Dipodomys | merriami | F | 35 | 45 | Rodent | DM |
| Chaetodipus | penicillatus | F | 20 | 15 | Rodent | PP |
| Dipodomys | merriami | M | 35 | 29 | Rodent | DM |
| Dipodomys | merriami | M | 35 | 39 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 43 | Rodent | DM |
| Dipodomys | merriami | M | 38 | 46 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 41 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 41 | Rodent | DM |
| Dipodomys | merriami | M | 38 | 40 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 45 | Rodent | DM |
| Dipodomys | merriami | F | 35 | 46 | Rodent | DM |
| Dipodomys | merriami | F | 35 | 40 | Rodent | DM |
| Dipodomys | merriami | F | 35 | 30 | Rodent | DM |
| Dipodomys | merriami | M | 35 | 39 | Rodent | DM |
| Dipodomys | merriami | M | 35 | 34 | Rodent | DM |
| Dipodomys | merriami | F | 37 | 42 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 42 | Rodent | DM |
| Perognathus | flavus | F | 13 | 8 | Rodent | PF |
| Dipodomys | merriami | F | 37 | 31 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 40 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 37 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 48 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 42 | Rodent | DM |
| Dipodomys | merriami | F | 39 | 45 | Rodent | DM |
| Chaetodipus | penicillatus | F | 21 | 16 | Rodent | PP |
| Dipodomys | merriami | F | 36 | 36 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 42 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 44 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 41 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 40 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 34 | Rodent | DM |
| Dipodomys | merriami | M | 33 | 40 | Rodent | DM |
| Dipodomys | merriami | M | 33 | 44 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 44 | Rodent | DM |
| Dipodomys | merriami | M | 34 | 36 | Rodent | DM |
| Dipodomys | merriami | M | 35 | 33 | Rodent | DM |
| Dipodomys | merriami | F | 37 | 46 | Rodent | DM |
| Dipodomys | merriami | F | 34 | 35 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 46 | Rodent | DM |
| Dipodomys | merriami | F | 33 | 37 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 34 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 45 | Rodent | DM |
| Perognathus | flavus | F | 15 | 7 | Rodent | PF |
| Dipodomys | merriami | M | 37 | 51 | Rodent | DM |
| Dipodomys | merriami | M | 35 | 39 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 29 | Rodent | DM |
| Dipodomys | merriami | F | 32 | 48 | Rodent | DM |
| Dipodomys | merriami | M | 38 | 46 | Rodent | DM |
| Dipodomys | merriami | F | 37 | 41 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 45 | Rodent | DM |
| Dipodomys | merriami | F | 35 | 42 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 53 | Rodent | DM |
| Dipodomys | merriami | F | 35 | 49 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 46 | Rodent | DM |
| Perognathus | flavus | F | 13 | 9 | Rodent | PF |
| Chaetodipus | penicillatus | F | 19 | 15 | Rodent | PP |
| Perognathus | flavus | M | 13 | 4 | Rodent | PF |
| Dipodomys | merriami | M | 36 | 48 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 51 | Rodent | DM |
| Dipodomys | merriami | M | 38 | 50 | Rodent | DM |
| Dipodomys | merriami | M | 35 | 44 | Rodent | DM |
| Dipodomys | merriami | M | 25 | 44 | Rodent | DM |
| Dipodomys | merriami | M | 35 | 45 | Rodent | DM |
| Dipodomys | merriami | F | 37 | 45 | Rodent | DM |
| Peromyscus | eremicus | M | 20 | 19 | Rodent | PE |
| Dipodomys | merriami | F | 38 | 44 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 42 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 39 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 47 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 42 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 49 | Rodent | DM |
| Dipodomys | merriami | M | 38 | 39 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 43 | Rodent | DM |
| Dipodomys | merriami | M | 35 | 50 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 41 | Rodent | DM |
| Dipodomys | merriami | M | 37 | 47 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 37 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 41 | Rodent | DM |
| Dipodomys | merriami | F | 36 | 36 | Rodent | DM |
| Dipodomys | merriami | M | 36 | 45 | Rodent | DM |
| Peromyscus | eremicus | M | 19 | 20 | Rodent | PE |
What if a species was included in the species dataset, but not in the measurement dataset?
Factor Variables
What is a factor variable?
In general, factors are used for:
- categorical variables with a fixed and known set of possible values.
- E.g.,
day_born= Sunday, Monday, Tuesday, …, Saturday
- displaying character vectors in non-alphabetical order.
Eras Tour
Let’s consider songs that Taylor Swift played on her Eras Tour. I have randomly selected 25 songs (and their albums) to consider.
Creating a Factor – Base R
[1] "Red" "Reputation" "Lover" "Midnights" "1989"
[6] "Fearless" "Reputation" "Folklore" "Midnights" "Evermore"
[11] "Evermore" "Lover" "Lover" "Red" "Reputation"
[16] "Reputation" "Speak Now" "Red" "Midnights" "Fearless"
[21] "1989" "Midnights" "Fearless" "Folklore" "Lover" [1] Red Reputation Lover Midnights 1989 Fearless
[7] Reputation Folklore Midnights Evermore Evermore Lover
[13] Lover Red Reputation Reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless Folklore
[25] Lover
9 Levels: 1989 Evermore Fearless Folklore Lover Midnights Red ... Speak NowCreating a Factor – Base R
When you create a factor variable from a vector…
- Every unique element in the vector becomes a level.
- The levels are ordered alphabetically.
- The elements are no longer displayed in quotes.
Creating a Factor – Base R
You can specify the order of the levels with the levels argument.
forcats
We use this package to…
turn character variables into factors.
make factors by discretizing numeric variables.
rename or reorder the levels of an existing factor.

forcats loads with tidyverse!
The packages forcats (“for categoricals”) helps wrangle categorical variables.
Creating a Factor – fct
With fct(), the levels are automatically ordered in the order of first appearance.
[1] Red Reputation Lover Midnights 1989 Fearless
[7] Reputation Folklore Midnights Evermore Evermore Lover
[13] Lover Red Reputation Reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless Folklore
[25] Lover
9 Levels: Red Reputation Lover Midnights 1989 Fearless Folklore ... Speak NowCreating a Factor
To change a column type to factor, you must wrap fct() in a mutate() call.
I am using pull() to display the outcome:
[1] Red Reputation Lover Midnights 1989 Fearless
[7] Reputation Folklore Midnights Evermore Evermore Lover
[13] Lover Red Reputation Reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless Folklore
[25] Lover
9 Levels: Red Reputation Lover Midnights 1989 Fearless Folklore ... Speak NowCreating a Factor – fct
You can still specify the order of the levels with level.
Creating a Factor – fct
You can also specify non-present levels.
Re-coding a Factor – fct_recode
Oops, we have a typo in some of our levels! We change existing levels with the syntax: "<new level>" = "<old level>".
eras_data |>
mutate(Album = fct_recode(.f = Album,
"folklore" = "Folklore",
"evermore" = "Evermore",
"reputation" = "Reputation")
)# A tibble: 25 × 2
Song Album
<chr> <fct>
1 22 Red
2 ...Ready for It? reputation
3 The Archer Lover
4 Bejeweled Midnights
5 Style 1989
6 You Belong With Me Fearless
7 Don't Blame Me reputation
8 illicit affairs folklore
9 Lavender Haze Midnights
10 marjorie evermore
# ℹ 15 more rowsRe-coding a Factor – case_when
We have similar functionality with the case_when() function…
eras_data |>
mutate(Album = case_when(Album == "Folklore" ~ "folklore",
Album == "Evermore" ~ "evermore",
Album == "Reputation" ~ "reputation",
.default = Album),
Album = fct(Album)) |>
pull(Album) [1] Red reputation Lover Midnights 1989 Fearless
[7] reputation folklore Midnights evermore evermore Lover
[13] Lover Red reputation reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless folklore
[25] Lover
9 Levels: Red reputation Lover Midnights 1989 Fearless folklore ... Speak NowCollapsing a Factor –fct_collapse
Collapse multiple existing levels of a factor with the syntax:
"<new level>" = c("<old level>", "<old level>", ...).
eras_data |>
mutate(Genre = fct_collapse(.f = Album,
"country pop" = c("Taylor Swift", "Fearless"),
"pop rock" = c("Speak Now", "Red"),
"electropop" = c("1989", "Reputation", "Lover"),
"folk pop" = c("Folklore", "Evermore"),
"alt-pop" = "Midnights")
) |>
slice_sample(n = 6)# A tibble: 6 × 3
Song Album Genre
<chr> <fct> <fct>
1 willow Evermore folk pop
2 You Belong With Me Fearless country pop
3 Lavender Haze Midnights alt-pop
4 We Are Never Ever Getting Back Together Red pop rock
5 illicit affairs Folklore folk pop
6 Look What You Made Me Do Reputation electropop Re-leveling a Factor –fct_relevel
Change the order of the levels of an existing factor.
Re-ordering Factors in ggplot2
The bars follow the default factor levels.

We can order factor levels to order the bar plot.
full_eras |>
mutate(Album = fct(Album,
levels = c("Fearless",
"Speak Now",
"Red",
"1989",
"Reputation",
"Lover",
"Folklore",
"Evermore",
"Midnights")
)
) |>
ggplot(mapping = aes(y = Album,
fill = Album)
) +
geom_bar() +
theme_minimal() +
theme(legend.position = "none") +
labs(x = "",
y = "",
title = "Number of Songs Played on the Eras Tour by Album")
Re-ordering Factors in ggplot2
The ridge plots follow the order of the factor levels.

Inside ggplot(), we can order factor levels by a summary value.

Re-ordering Factors in ggplot2
The legend follows the order of the factor levels.
full_eras |>
filter(!Album %in% c("1989","Fearless")) |>
group_by(Album, Single) |>
summarise(avg_len = mean(Length)) |>
ggplot(mapping = aes(x = Single,
y = avg_len,
color = Album)) +
geom_point(size = 1.5) +
geom_line() +
theme_minimal() +
scale_x_continuous(breaks = c(0,1),
labels = c("No", "Yes")
) +
labs(y = "",
title = "Are Taylor Swift's Singles Shorter?",
color = "Album")
Inside ggplot(), we can order factor levels by the \(y\) values associated with the largest \(x\) values.
full_eras |>
filter(!Album %in% c("1989","Fearless")) |>
group_by(Album, Single) |>
summarise(avg_len = mean(Length)) |>
ggplot(mapping = aes(x = Single,
y = avg_len,
color = fct_reorder2(.f = Album,
.x = Single,
.y = avg_len)
)
) +
geom_point(size = 1.5) +
geom_line() +
theme_minimal() +
scale_x_continuous(breaks = c(0,1),
labels = c("No", "Yes")
) +
labs(y = "",
title = "Are Taylor Swift's Singles Shorter?",
color = "Album")
Lab 4: Childcare Costs in California
ChatGPT to the Rescue!
To do…
- Lab 4: Childcare Costs in California
- Due Sunday (10/20) at 11:59pm
- Read Chapter 5: Strings + Dates
- Check-in 5.1 due Tuesday (10/22) at 12:10pm
- Check-in 5.2 due Thursday (10/24) at 12:10pm
